Research
Main content
Intracellular biomacromolecules collectively self-organize into subcellular structures and dynamic condensations essential in physiological processes. We are interested in understanding how the collective behaviors of these biomacromolecule can give rise to the biological phenomena, such as Liquid-liquid phase separation (LLPS) and self assembly. Particularly, given the far-out-of-equilibrium nature of living systems, we are interested in how internally driven components and chemical reactions can give rise to new phase behaviors.
Representative results are given as follows:
Motion transition of active filaments
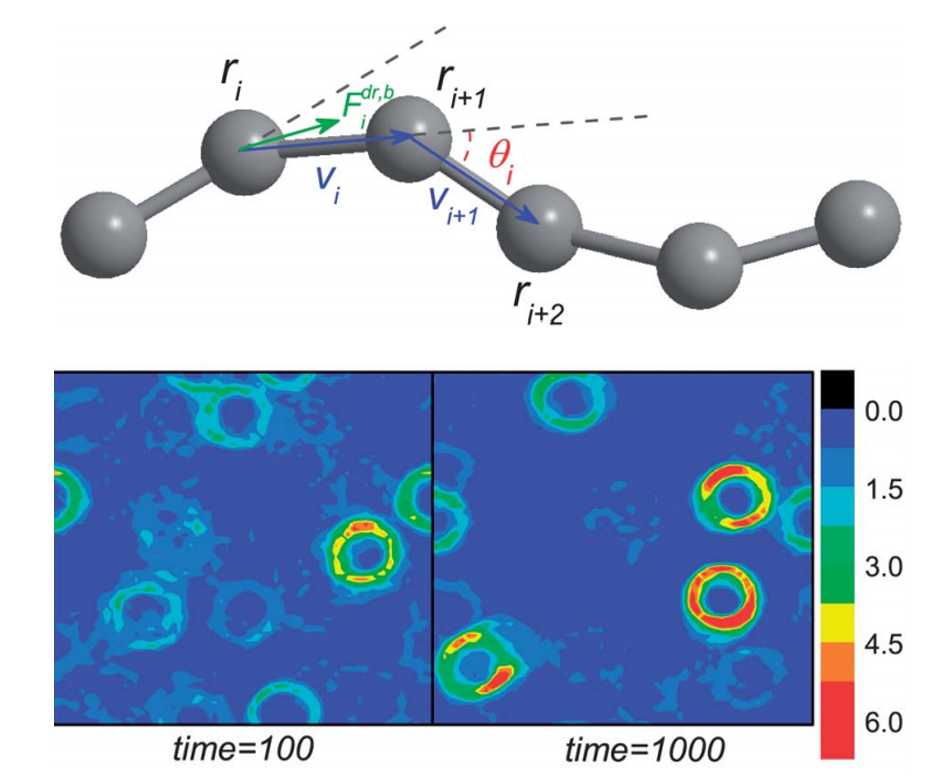
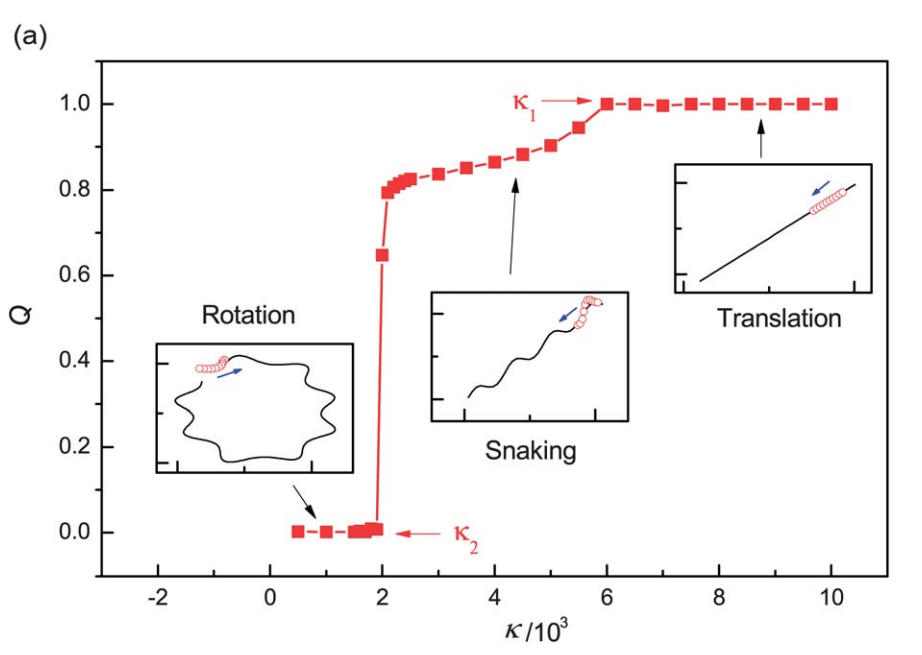
Active filaments can self-organize into fascinating coherently moving structures, such as propagating density waves and vortexes. We find that the filament can show three distinct types of motion, namely, translation, snaking and rotation, with the variation of the rigidity or active force. Of particular interest, we find that HI is not necessary for the rotation or snaking motion, but can enlarge remarkably the parameter regions in which they can occur. Our findings provide new insights into the subtle role of HI in the formation of collective structures in active systems
Soft Matter, 2014, 10, 1012–1017
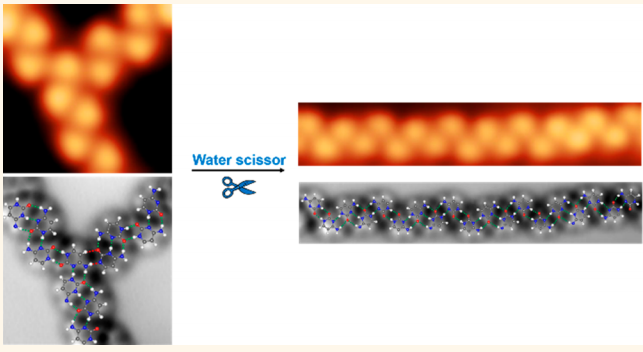
Hydrogen-Bond Regulation on Cytosine Dimer Assemblies
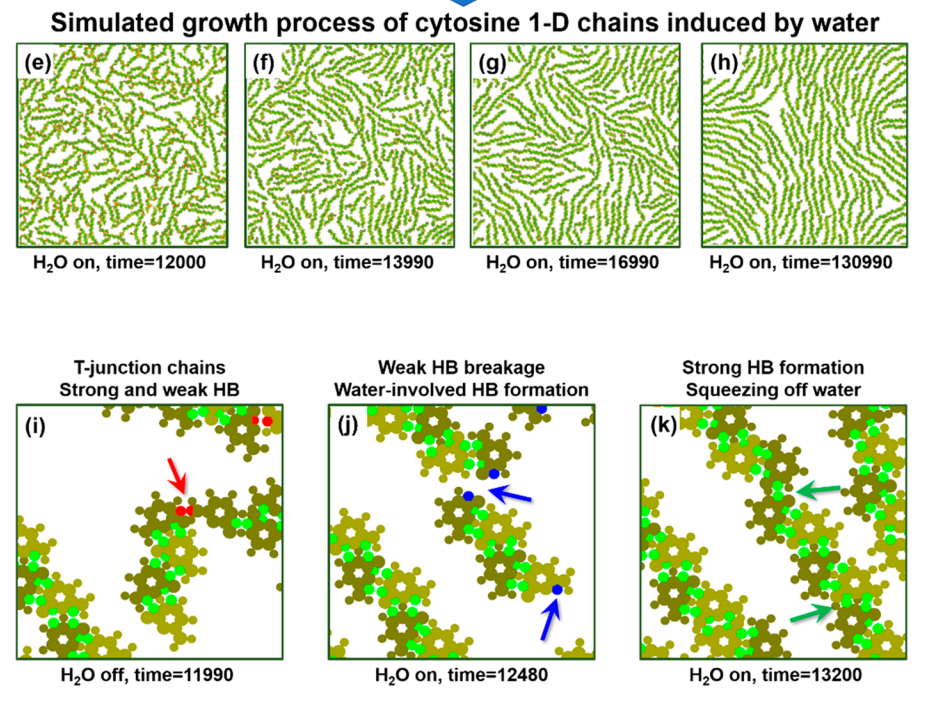
The solvation of one of the DNA bases, cytosine, whose glassy-state network formed on Au(111) contains diverse types of hydrogen-bonded dimer configurations with hierarchical strengths are investigated. Upon water exposure, a global structural transformation from interwoven chain segments to extended chains Density functional theory calculation and coarse-grained molecular dynamics simulation indicate that water molecules selectively break the weak-hydrogen-bonded dimers at T-junctions, while the stable ones within chains remain intact. Such an intriguing transformation cannot be realized by thermal annealing, indicating the dynamic nature o of water molecules in the regulation of hydrogen bonds in a catalytic manner.